Another typical aplication for solids, is to use them as molecules and clusters carriers.
A metal cathalytic cluster is active due to its bond insaturations, this property makes it
very reactive, so it can bond to another cluster, and so on, to form a solid, eliminating
its reactivity. Also surfaces can improve the molecule-cluster properties, by changing its
electronic and geometric structure. It is important to study the interactions between them.
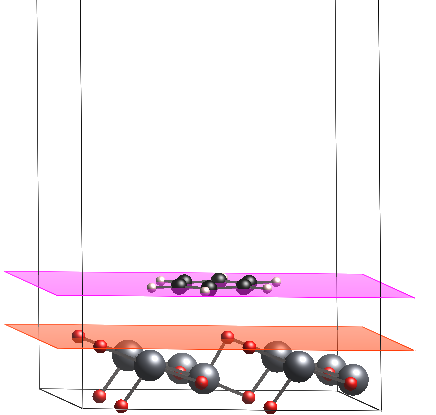
Figure 1. Interaction between Benzene molecule and Titanium Dioxide surface [110], the purple plane indicates the benzene surface and red one indicates the TiO2 surface. Builded on it on Virtual NanoLab.
Relaxation
Due to the size of the system, this type of calculation will take too long, to make a scanning of where the molecule has more probability to be adsorbed by the surface, we can make a first aproximation by freezing the atom surfaces.
POSCAR constructionLets build the benzene-anatase system, there are three ways:
- Convert the anatase POSCAR from direct to cartesian coordinates
- Open the anatase poscar file and add the benzene cartesian coordinates.
- Visualize it with VESTA and translate them manually, until you get the desire possition.
- Open anatase and benzene .cif files in MS-modelling.
- Visualize the benzene molecule, they have the same box size, copy to clipboard the benzene molecule, (select all molecule and press Ctrl+c).
- Visualize the anatase structure. Paste the benzene molecule (Ctrl+v).
- Translate benzene molecule to the desired possition.
possition.
- Excecute vnl on the directory where you have both cif files.
- Use the builder tool.
- Load both .cif files.
- On the right menubar click on +Bulk Tools> Mergecells.
- Drag the anatase structure to the black zone, then drag the benzene cell.
- Click on merge
- Translate the benzene until the desired possition.
To froze the atoms we have to add a new line called Selective Dynamics, and to indicate which atoms and which coordinates we want to freeze.
Anatase + Benzene
1.0
11.1524000168 0.0000000000 0.0000000000
-2.5919744258 7.1481640748 0.0000000000
0.0000000000 0.0000000000 17.9026985168
Ti O C H
8 16 6 6
Selective dynamics
Direct
0.473363012 0.463707000 0.045630001 F F F
0.239115998 0.096606001 0.086956002 F F F
0.973362982 0.463707000 0.045630001 F F F
0.739116013 0.096606001 0.086956002 F F F
0.473363012 0.963707983 0.045630001 F F F
0.239115998 0.596605957 0.086956002 F F F
0.973362982 0.963707983 0.045630001 F F F
0.739116013 0.596605957 0.086956002 F F F
0.283683002 0.368804991 0.082051001 F F F
0.060326997 0.007208000 0.129732996 F F F
0.425368994 0.189715996 0.048241001 F F F
0.134815007 0.044450000 0.003833000 F F F
0.783683002 0.368804991 0.082051001 F F F
0.560326993 0.007208000 0.129732996 F F F
0.925369024 0.189715996 0.048241001 F F F
0.634814978 0.044450000 0.003833000 F F F
0.283683002 0.868804991 0.082051001 F F F
0.060326993 0.507207990 0.129732996 F F F
0.425368994 0.689715981 0.048241001 F F F
0.134815007 0.544449985 0.003833000 F F F
0.783683002 0.868804991 0.082051001 F F F
0.560326993 0.507207990 0.129732996 F F F
0.925369024 0.689715981 0.048241001 F F F
0.634814978 0.544449985 0.003833000 F F F
0.559710026 0.692732990 0.239644021 T T T
0.645358026 0.595155001 0.239644021 T T T
0.600000024 0.399999976 0.239644021 T T T
0.468995005 0.302421987 0.239644021 T T T
0.383347005 0.399999976 0.239644021 T T T
0.428705007 0.595155001 0.239644021 T T T
0.595466018 0.846576989 0.239644021 T T T
0.748636007 0.672083974 0.239644021 T T T
0.667518973 0.323071003 0.239644021 T T T
0.433238983 0.148578018 0.239644021 T T T
0.280068994 0.323071003 0.239644021 T T T
0.361185998 0.672083974 0.239644021 T T T
INCAR preparation
In this example we are going to relax the ion possition in the cell, because symmetry is broken and the number of atoms increase, VASP will take more ion relaxation steps, so be patient. The INCAR file must have the followin directives.
- ISTART= 0 New Job.- ISIF = 2 Ions relaxation.
- IBRION= 2 DFT Conjugate Gradient.
- NSW = 25 Maximum Optimization steps.
- EDIFF & EDIFFG Convergence Criteria. - ICHARG=2 Initial charge distribution are the atomic charges.
# Anatase + Benzene interaction #=========================================== # General Setup System = Anatase_benz # Calculation Title PREC = NORMAL # Options: Normal, Medium, High, Low ENCUT = 400 # Kinetic Energy Cutoff in eV ISTART = 0 # Job: 0-new 1-cont 2-samecut ICHARG = 2 # initial charge density: 1-file 2-atom 10-cons 11-DOS ISPIN = 1 # Spin Polarize: 1-No 2-Yes # Electronic Relaxation (SCF) NELM = 60 # Max Number of Elec Self Cons Steps NELMIN = 2 # Min Number of ESC steps NELMDL = 10 # Number of non-SC at the beginning EDIFF = 1.0E-04 # Stopping criteria for ESC LREAL = .TRUE. # Real space projection IALGO = 48 # Electronic algorithm minimization VOSKOWN = 1 # 1- uses VWN exact correlation ADDGRID = .TRUE. # Improve the grid accuracy # Ionic Relaxation EDIFFG = 1.0E-02 # Stopping criteria for ionic self cons steps NSW = 55 # Max Number of ISC steps: 0- Single Point IBRION = 2 # Ionic Relaxation Method: 0-MD 1-qNewton-RaphsonElectronic 2-CG ISIF = 2 # Stress and Relaxation: 2-Ion 3-cell+ion ADDGRID = .TRUE. # Improve the grid accuracy SIGMA = 0.10 # Insulators/semiconductors=0.1 metals=0.05 ISMEAR = 0 # Partial Occupancies for each Orbital # -5 DOS, -2 from file, -1 Fermi Smear, 0 Gaussian Smear # Parallelization NPAR=8 NCORE=8
POTCAR
We have to combine the potential files of anatase and benzene. To do that copy the POTCAR files into your work directory with diferent name. Potential files are in the directory where you install VASP. Finally just concatenate both files into a new file called POTCAR. Remember conserve the order that we impose in the POSCAR.
[user@machine WRKDIR]$ cat POTCAR_TiO2 POTCAR_Benzene >> POTCAR
KPOINTS file
We are going to generate an automatic K-mesh with 36 K-points distributed in all directions.
Automatic Mesh # Generates Automatically the K-mesh 0 Monkhorst-pack # Automatic Distribution of the mesh 6 6 1
Due that there is no periodicity on the Z axis, there is only one KPOINT on Z.
Job administrator
If you have a queue administrator you have to set the queue file. FamaLab has torque as default job manager. Please paste the queue file .pbs or .sge into your work directory.
The file has as main directives the jobname, the number of cpu you are requesting, the time is going to take your calculation, and which of the vasp programs you will use.
#PBS -N name_identifier #PBS -o vasp.out #PBS -j oe #PBS -e vasp.err #PBS -l walltime=8760:00:00 #PBS -l nodes=1:ppn=8 #PBS -q batch #Elegir la version de vasp: VASP=vasp_cd #Volume=vasp_complex, supercell=vasp_cd, molecules=vasp_gamma
The variables you can modify are marked in red:
You can download the build files here:
INCAR
POTCAR
KPOINTS
POSCAR